Martins J. P. A., Melo E. B., Ferreira M. M. C., "2D-QSAR study of antimutagenic flavonoids using Ordered Predictors Selection (OPS)". Porto de Galinhas, PE, Brazil, 09-13/11/2008: The 4th Brazilian Symposium on Medicinal Chemistry (BrazMedChem2008): Systems Chemical Biology, CD-ROM Online, (2008) No. 11. Poster R0020-1.
Brazilian Chemical Society (SBQ). Division of Medicinal Chemistry. 4th Brazilian Symposium on Medicinal Chemistry
2D-QSAR study of antimutagenic flavonoids using Ordered Predictors Selection (OPS)
Martins, J. P. A.1; Melo, E. B.1,2; Ferreira, M. M. C.1 *joaopauloam@gmail.com
1IQ/Universidade Estadual de Campinas; 2Farmácia/Universidade Estadual do Oeste do Paraná.
Keywords: QSAR; PLS; variable
selection; antimutagenic effect; flavonoids; 3-NFA.
Introduction
Nitroarenes, like the nitrofluoranthene
(3-NFA), are generated by reaction of nitrogen oxides (NOx) with polycyclic
aromatic hydrocarbons during incomplete combustion of organic materials.
These compounds present mutagenic activities in bacterial and mammalian
test systems and are associated with some types of cancer. However, carcinogenicity
and mutagenicity of chemicals may be modulated by other chemicals. For
example, it is known that the flavonoids found in food possess protective
properties [1].
In this study was conducted
a 2D-QSAR study with twenty selected flavonoids among that studied by Endeharder
and Tang [1] that presented inhibition of mutagenic activity promoted by
3-NFA in Salmonella typhimurium TA98.
A total of 1221 descriptors
were calculated with the programs Gaussian 03 (AM1), PClient (www.vcclab.org)
and DRAGONWEB. A cut off was performed and 649 descriptors with correlation
coefficient lower than 0.4 with the biological activity were eliminated.
In a further step, a variable selection was carried out using the Ordered
Predictors Selection (OPS) algorithm [2]. The OPS algorithm searches for
the best models until 5 latent variables (LV), obey the criterion Nsamples/NLV
> 3. The best model was submitted to a series of internal (cross-validation,
y-randomization and leave-N-out) and external validation in order to confirm
its reliability, robustness and check for chance correlation.
Results and Discussion
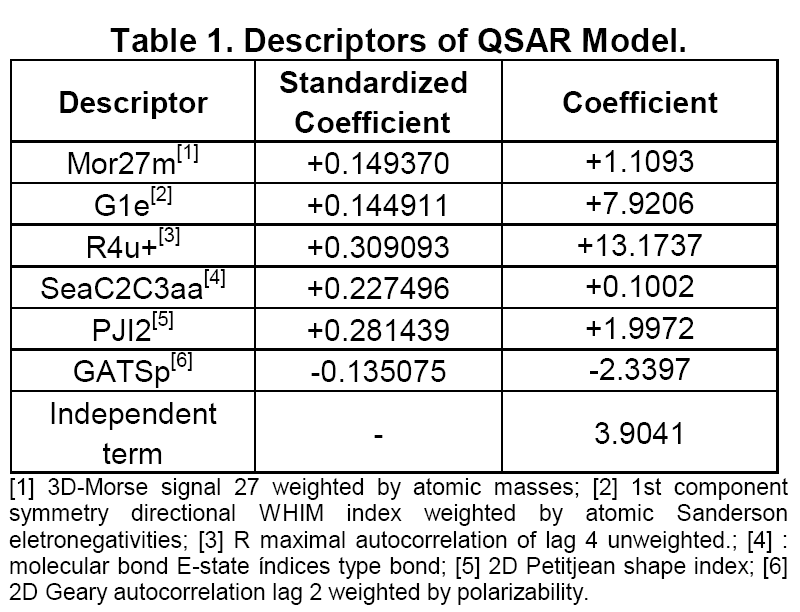
The OPS algorithm has suggested 330 different models and the best model was chosen based on the minimum Spress [3], resulting in a PLS model with 2 latent variables, 6 descriptors, and 70.78% of total information. One outlier was removed based on its Leverage and Studentized residual and the final model had 19 samples. The model (Table 1) presented good statistical results (R2 = 0.7708, SEC = 0.3162, PRESScal = 1.5997, Q2LOO = 0.5858, SEV = 0.3900, PRESSval = 2.8905 and F95% = 28.692 for Fcritical = 3.633). All of selected descriptors were topological. However G1e, SeaC2C3aa and GATSp indicate effects related to electronegativity and polarizability that should be taken into account.
In the leave-N-out validation
analysis (N = 1 to 5) the average Q2 was
0.5649±0.024 (max: 0.5983; min: 0.5305), showing small oscillation
in the results and confirming the robustness of the model. A y-randomization
test was also performed (R2ís < 0.3
and Q2ís < 0.0), showing no chance correlation.
After all processes of internal validation, a test set with 5 samples were
selected among the 19 samples of the full set and new models were built
(similar statistics from Table 1). The external validation presented the
following results: Q2pred =
0.9389, SEP = 0.2684, PRESSpred = 0.3601
and average error of prediction 3.8012%. These results show that the model
is also good to perform predictions.
Conclusions
The model obtained by OPS
algorithm presented good statistical results, robustness and a great ability
of prediction. Thus, it can be used as a guide to the synthesis of new
flavonoids with better ability of chemoprevention regarding the antimutagenic
effects caused by nitroarenes.
Acknowledgements
FAPESP, CNPQ and Universidade
Estadual do Oeste do Paraná/Campus Cascavel.
1 Edenharder, R.; Tang, X. Food
Chem. Toxicol., 1997, 35, 357.
2 Teófilo, R.F.; Martins,
J.P.A.; Ferreira, M.M.C. J. Chem. 2008, xx.
3 Kubinyi, H. 3D-QSAR in drug
design. Theory methods anapplications. Kluwer academic publishers. 2000,
538.
4th Brazilian Symposium on Medicinal Chemistry - BrazMedChem2008