Pereira F. S., Barbosa E. G., Pasqualoto K. F. M., Ferreira M. M. C., "4D-LQTAqsar analysis of a set of antimalarial compounds". Porto de Galinhas, PE, Brazil, 09-13/11/2008: The 4th Brazilian Symposium on Medicinal Chemistry (BrazMedChem2008): Systems Chemical Biology, CD-ROM Online, (2008) No. 81. Poster R0223-1.
Brazilian Chemical Society (SBQ). Division of Medicinal Chemistry. 4th Brazilian Symposium on Medicinal Chemistry
4D-LQTAqsar ANALYSIS OF A SET OF ANTIMALARIAL COMPOUNDS
Pereira, F. S.; Barbosa, E. G.; Pasqualoto, K. F. M.; Ferreira, M. M. C.
Instituto de Química,
Universidade Estadual de Campinas, CP 6154, 13084-971 Campinas - SP, Brasil
marcia@iqm.unicamp.br
Keywords: QSAR, ARTEMISININ,
PLS, LQTAgrid program
Introduction
According to the World Health
Organization (WHO)1 the malaria occurs
in over 100 countries, and more than 40% of the world's population is living
at risk. The conventional chemotherapeutic agents have reduced its effectiveness,
because the increasing of malaria parasites resistance to the existing
drugs, causing a real threat to the control of malaria. Artemisinin and
its derivatives emerged as a new class of antimalarials, which are effective
against drug-resistant strains of Plasmodium falciparum.
This study aims the construction
of QSAR models of a set of thirty-three artemisinin derivatives, including
artemisinin as the lead drug, using partial least squares (PLS) regression.
All biological data used in this work were expressed as logarithm of relative
activity, logRA,2 calculated considering
equation 1.
log RA = log
[(IC50 of artemisinin/ IC50
of the analogue) x (MW of the analogue/MW of the artemisinin)]
(eq.1)
where: MW =
molecular weight.
Artemisinin crystal structure
retrieved from Cambridge structural Database, reference code QNGHSU033
(crystallography R factor 3.60%), was used as starting geometry to build
all the ligands. The complete geometry optimizations of artemisinin and
its derivatives were carried out employing MM+ force field, AM1 semiempirical
method, ab initio HF/3-21G and HF/6-31G methods, and DFT using B3LYP/6-311++G**
as basis set. The Gaussian, HyperChem, and 98W programs were used for molecular
modeling calculations and PIROUETTE4 package
was employed to carry out the chemometric analysis.
All thermodynamic descriptors
(van der Waals and electrostatic energy contributions) were calculated
employing the LQTAgrid program, developed in our group (Barbosa, EG; Martins,
JPA; Pasqualoto, KFM; Ferreira, MMC), using NH3+ as a probe, and considering
the conformational profile (PC) of each ligand. The formalism employed
in this work combines the advantages of the methods, CoMFA and independent-receptor
4D-QSAR.
Results and Discussion
The best QSAR model (N =
33) presented the following statistical parameters values: q2
= 0.59; r2 = 0.77; SEV = 0.37; SEC = 0.3;
and, SEP = 0.28, using 4 latent variables. The resulting QSAR model was
validated applying Y-randomization and leave-N-out (N = 1 to 10)
methodologies.
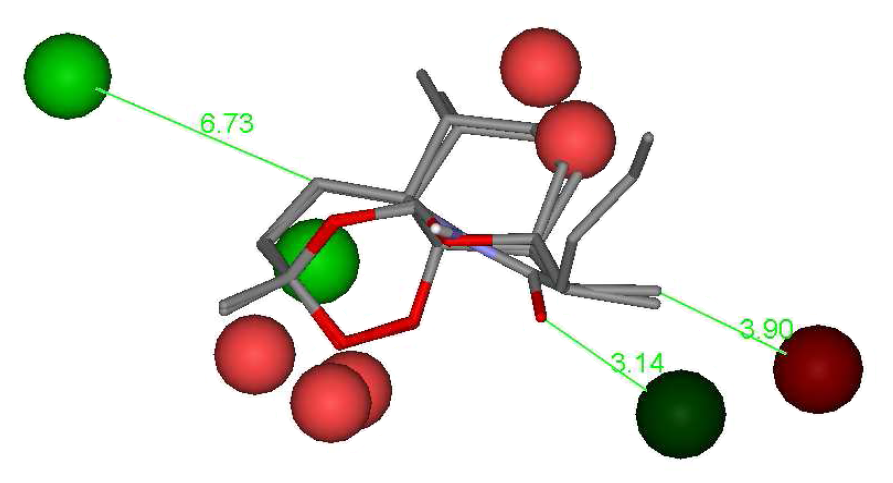
The descriptors selected
in the best QSAR models can be graphically visualized (hot spots)
and are presented in Fig. 1. Favorable and unfavorable energy contributions
to the biological activity are shown as green (dark and light) and red
(dark and light) spheres, respectively. Figure 1 shows the graphical visualization
of the 4D descriptors selected in the best QSAR model. Lennard-Jones descriptors
are presented as light color spheres and Coulombic descriptors as dark
spheres, respectively.
Figure 1. Superposition
of two artemisinin derivatives and the 4D-descriptors selected in the best
QSAR model.
Conclusions
The methodology applied in
this study generated a QSAR model having a reasonable internal and external
predictability. These findings can be helpful in the design of new antimalarial
agents.
Acknowledgements
The authors are grateful
to CNPq, CAPES, and FAPESP for financial support.
1 http://www.who.int/malaria/
Accessed in 10/04/2008.
2 Avery, M. A.;Alvim-Gaston,
M.; Rodrigues, C.R.; Barreiro, E.J.; Cohen, F.E.; Sabnis, Y.A.;Woolfrey,J.R.,
J.
Med. Chem. 2002, 45, 292.
3 Lisgarten, J. N.; Potter,
B. S.; Bantuzeko, C.; Palmer, R. A., J. Chem. Crystallogr. 1998,
28,
539.
4 INFOMETRIX, INC; PIROUETTE
3.01, Woodinville, WA, 2001.
4th Brazilian Symposium on Medicinal Chemistry - BrazMedChem2008