Teófilo R. F., Ataide J. P., Ferreira M. M. C., "Study of the computational performance of PLS algorithms using experimental design". Lappeenranta, Finland, 11-15/07/2007: 10th Scandinavian Symposium on Chemometrics (SSC10), Book of Abstracts, (2007) 97. Poster PO35.
10th Scandinavian Symposium on ChemometricsPO 35
Study of the computational
performance of PLS algorithms
using
experimental design
Reinaldo Francisco Teófilo, João Paulo
Ataide Martins, Márcia Miguel Castro Ferreira
Laboratório de
Quimiometria Teórica e
Aplicada - Instituto de
Química - Universidade
Estadual de Campinas
_____________________________________________________________________________________
1. Introduction
Among the multivariate calibration methods,
Partial Least Squares (PLS) regression have been chosen
due to relevant advantages over other methods.
In distinct applications of multivariate calibration, data
matrices can be very large,
e.g. in 3D-QSAR, data mining, near infrared
spectroscopy (NIR), and
calculation time is a factor which cannot be neglected.
Thus, a fast PLS algorithm is very important
in
this situation, since time can be saved during model
building.
The purpose of this work is to compare, using
experimental design, five PLS algorithms available in the
literature with respect to their
computational time when doing leave-one-out cross-validation. They
are:
classical NIPALS (NIPALS) [1],
modified NIPALS (NIPALSy) [2], Kernel (Kernel)
[3], SIMPLS
(SIMPLS) [4], the bidiagonalization algorithm
(PLSBi) [5]. Different dimensions matrices were
tested
aiming to show which algorithm is the best in each situation.
2. Experimental
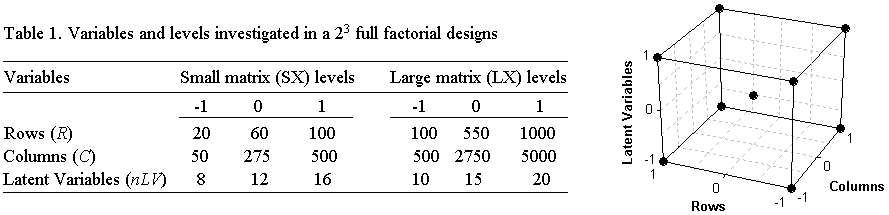
Two full factorial designs, 23,
with center point in triplicate were carried out considering two types
of data
matrix X: small (SX) and large
(LX). The response investigated in the
experimental design was the
running time of the algorithm during cross-validation.
Table 1 shows the variables investigated
and the
levels studied for each data set.
The independent variables X and
dependent y were generated with random
numbers and matrix
dimensions as described in Table
1. The same leave-one-out cross-validation
was applied for all
algorithms with the number of latent variables defined
in Table 1.
3. Results and discussion
It was observed that row and column numbers are, as expected,
the major factors influencing the response
time. Except for Kernel,
the columns were more important that rows for
SX and the opposite was
observed for LX. The nLV variable
showed to be of little significance in running time compared
to X
dimensions.
It is possible to conclude that
between PLSBi and SIMPLS a small differnce
in performance was
observed but they were more efficient
than the others, mainly for large data sets.
Among NIPALS,
NIPLALSy and Kernel, the performance in descending
order was: NIPALSy, NIPALS and Kernel. It is
important to emphasize that all algorithms present the
same prediction results for all data set tested.
_____________________________________________________________________________________
References
[1] Haaland, D. M. & Thomas, E. V., Anal. Chem. 60,
1988, 1193-1202.
[2] Dayal, B. S. & MacGregor, J. F., J. Chemometr.
11, 1997, 73-85.
[3] Lindgren, F., Geladi, P. & Wold, S., J. Chemometr.
7, 1993, 45-59.
[4] de Jong, S., Chemometrics Intell. Lab. Syst. 18,
1993, 251-263.
[5] Wu, W. & Manne, R., Chemometrics Intell. Lab.
Syst. 51, 2000, 145-161.
97