Kiralj R., Ferreira M. M. C., "Theoretical Study on Some b-Lactams as Substrates of the Bacterial Multidrug Resistance AcrB Pump". Caxambu, MG, 23-26/11/2003: 12° Simpósio Brasileiro de Química Teórica (XII SBQT) [12th Brazilian Symposium of Theoretical Chemistry], Livro de Resumos [Book of Abstracts], (2003) P149. Poster P149.
|
|
THEORETICAL
STUDY ON SOME b-LACTAMS
AS
SUBSTRATES
OF THE BACTERIAL MULTIDRUG RESISTANCE
ACRB
PUMP
Rudolf
Kiralj (PQ), Márcia M. C. Ferreira (PQ)
marcia@iqm.unicamp.br
Instituto
de Química - UNICAMP, Campinas SP, 13084-971 Brazil.
Palavras-chave: b-lactam conformation, molecular mechanics, chemometrics
AcrB pump is a constitutive part of
the most important multidrug efflux system of
gram-negative bacteria,
which excretes a variety of compounds from bacterial
cytoplasm and
periplasm directly
to the cell exterior [1].
b-Lactam
antibiotics, the most widely used
antibacterial drugs,
are also substrates of this pump system. This fact
seriously increases the
problems in the treatment
of infectous diseases.
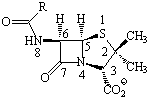
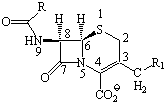
Montecarlo conformational search was performed
for 16 b-lactams
(penicillins and
cephalosporins, Scheme)
and their geometry was optimized
at semi-empirical PM3 and
molecular mechanics MMFF94
level. Various geometric, electronic, hydrogen bond, topological
and lipophilicity
molecular descriptors were calculated. Chemometric
analysis through Partial
Least Squares (PLS)
regression models was performed for prediction of b-lactam
efflux rates
(minimal inhibitor concentration,
MIC) caused by three distinct Salmonella typhimurium
strains
[1]. Docking
of selected drugs to the pore recognition site (PRS) of the
AcrB crystal structure
from Escherichi
coli [2] at MMFF94 level was also performed.
It has been already confirmed that lipophilicity and charges in b-lactam
molecules are
important in their excretion
by all bacterial strains, and that these drugs were classified as
good,
moderately good to poor,
and bad AcrB substrates [3]. In this work, parsimonius
PLS models
were obtained (Q2
> 0.67, R2 > 0.79). Several molecular
descriptors were selected: lipophilicity
(two log of octanol/water
partitition coefficients in linear and quadratic form; surface
fraction of
hydrophobic
carbon atoms), electronic (number fraction
of heteroatoms; second order
hyperpolarizability;
average atomic number in R and R1
substituents) and hydrogen bond
(average number
of hydrogen bonds) descriptors.
Both the experimental and predicted
activities pointed out
that liphophilic and amphiphilic drugs are
better AcrB substrates than
hydrophilic molecules.
Steric (size/shape) molecular descriptors
did not exibit significant
correlation with biological
activities, what agrees with the docking
studies where the pore
channel chains suffered
mostly minor conformational changes. Small but
visible and regular
changes occur in relative
position of the highly polar residues at the PRS. The
conformational
changes during the
docking are remarkably visible in docked
drugs, due to hydrophobic-
hydrophobic,
polar-polar and hydrogen bond interactions between
the drugs and the PRS.
Thus, the conformational
changes in the side chains R and R1,
and also in the b-lactam
ring
provoke variations
in electronic properties, and enable the
drug binding to the PRS via
hydrogen bonds and polar-polar
interactions. It can be concluded that electronic structure of the
drugs, directly and indirectly,
rather than their molecular geometry, determines the drug –
AcrB
pump interactions.
(FAPESP)


Scheme. Penicillins and cephalosporins
[1] H. Nikaido et al., J.
Bacteriol. 180 (1998) 4686.
[2] S. Murakami et al.,
Nature 419 (2002) 914.
[3] M. M. C. Ferreira, Kiralj,
R., J. Bacteriol., submitted.